SCMS: Insights into xenobiotic uptake
Pharmaceutical residues introduced to agricultural environments as a result of irrigation with reclaimed wastewater have been shown to have toxicological effects on crops and to accumulate in plant tissues. A high-resolution assessment of plant uptake and the distribution of priority compounds was accomplished through the application of single-cell mass spectrometry (SCMS).
SCMS was used to sample and analyse the contents of single plant cells exposed to exogenously applied pharmaceutical compounds. Parent compounds and transformation products were identified, including bioactive metabolites, some of which were previously undocumented in plant uptake studies thus providing an insight into the fate of pharmaceuticals once inside a cell following plant uptake.
SCMS was used to sample and analyse the contents of single plant cells exposed to exogenously applied pharmaceutical compounds. Parent compounds and transformation products were identified, including bioactive metabolites, some of which were previously undocumented in plant uptake studies thus providing an insight into the fate of pharmaceuticals once inside a cell following plant uptake.
|
Pharmaceuticals in the environment
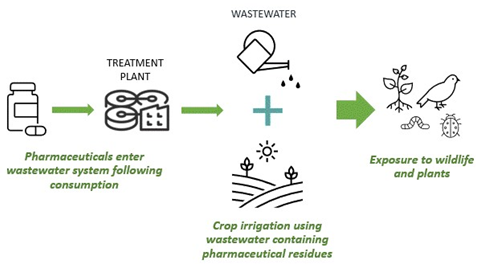
Reclaimed wastewater is increasingly used for irrigation of arable crops to alleviate water scarcity pressures. However, wastewater is a known reservoir for pharmaceutical residues which are subsequently introduced into the soil-plant system (Figure 1). Pharmaceutical uptake in plants has thus been increasingly documented as an unintended side effect of this irrigation technique. Uptake and accumulation of several pharmaceutical groups has now been documented in the tissues of edible plant species (1). Not only has this frequently been associated with growth and nutrient abnormalities in exposed plants, but recent studies have demonstrated that consumption of crops irrigated with pharmaceutical contaminated wastewater provides a direct human and wildlife exposure pathway via the food chain (2,3). |
SCMS and current research
Although numerous studies have investigated the fate of pharmaceuticals following plant uptake, these findings exclusively use techniques which analyse tissues in bulk and isolate cells from their in situ environments (1).
The results of these studies are therefore representative of a perturbed state, describing analytes as they occur under altered conditions, compared with those in undisturbed plant tissues (4). These studies also lack the resolution required to identify if pharmaceutical xenobiotics are able to enter single plant cells or elucidate the mechanisms and pathways occurring within cells to transform, fix and eliminate pharmaceuticals following uptake. Existing research also often uses a targeted approach, focusing almost exclusively on the detection of parent compounds in plant tissues, without considering potential transformation products that may have formed following plant uptake and thus are relevant when considering food chain exposure.
With this in mind, a novel combination of experimental approaches was developed at the University of York in collaboration with the Centre of Excellence in Mass Spectrometry. Exogenously applied pharmaceutical compounds were screened and analysed in single plant cells using SCMS. SCMS is a technique by which the contents of individual cells can be rapidly analysed in situ using high resolution mass spectrometry (5,6). Using SCMS, it is possible to detect and identify directly molecules in complex biological samples through analysis of a detailed spectrum of molecular peaks derived from molecular species that reveal the sample composition. Our study aimed to determine whether unchanged pharmaceutical residues and transformation products could accumulate in detectable concentrations within single plant cells.
Although numerous studies have investigated the fate of pharmaceuticals following plant uptake, these findings exclusively use techniques which analyse tissues in bulk and isolate cells from their in situ environments (1).
The results of these studies are therefore representative of a perturbed state, describing analytes as they occur under altered conditions, compared with those in undisturbed plant tissues (4). These studies also lack the resolution required to identify if pharmaceutical xenobiotics are able to enter single plant cells or elucidate the mechanisms and pathways occurring within cells to transform, fix and eliminate pharmaceuticals following uptake. Existing research also often uses a targeted approach, focusing almost exclusively on the detection of parent compounds in plant tissues, without considering potential transformation products that may have formed following plant uptake and thus are relevant when considering food chain exposure.
With this in mind, a novel combination of experimental approaches was developed at the University of York in collaboration with the Centre of Excellence in Mass Spectrometry. Exogenously applied pharmaceutical compounds were screened and analysed in single plant cells using SCMS. SCMS is a technique by which the contents of individual cells can be rapidly analysed in situ using high resolution mass spectrometry (5,6). Using SCMS, it is possible to detect and identify directly molecules in complex biological samples through analysis of a detailed spectrum of molecular peaks derived from molecular species that reveal the sample composition. Our study aimed to determine whether unchanged pharmaceutical residues and transformation products could accumulate in detectable concentrations within single plant cells.
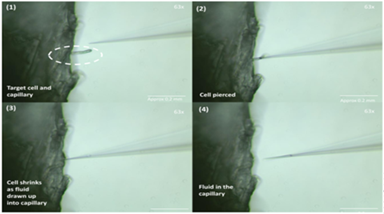 Figure 2. Image sequence showing the process of sampling single cellular material from a root hair cell of an Arabidopsis seedling.
Figure 2. Image sequence showing the process of sampling single cellular material from a root hair cell of an Arabidopsis seedling.
Experimental approach
Seedlings were hydroponically exposed to high concentration pharmaceutical solutions containing either the antiepileptic compound levetiracetam, or one of four antibiotic compounds (metronidazole, clarithromycin, sulfadiazine and lincomycin).
Fine cross sections were taken from the stem of the seedlings and visualised under a stereo microscope equipped with a micromanipulator and suction device. Hand-pulled, sharply tapering borosilicate capillaries were then used to puncture the cell wall of single cells from each of the cross sections (Figure 2). Under negative pressure conditions, the contents of each cell were drawn up into the sampling capillary and flash frozen at -80°C. The collected sample volumes were very small as a consequence of sampling single cells (<low μL). The cellular contents were then sprayed directly from the sampling capillary into a high resolution, high mass accuracy Orbitrap mass spectrometer using a static nanospray ionisation source. This technique provides a means of analysis that uses a soft ionisation technique to convey analytes into the mass spectrometer, minimising fragmentation of constituent molecules, making it suitable for analysis of complex biological samples. The mass spectra from each sample were then screened for the parent compounds and potential pharmaceutical metabolites and conjugates.
Seedlings were hydroponically exposed to high concentration pharmaceutical solutions containing either the antiepileptic compound levetiracetam, or one of four antibiotic compounds (metronidazole, clarithromycin, sulfadiazine and lincomycin).
Fine cross sections were taken from the stem of the seedlings and visualised under a stereo microscope equipped with a micromanipulator and suction device. Hand-pulled, sharply tapering borosilicate capillaries were then used to puncture the cell wall of single cells from each of the cross sections (Figure 2). Under negative pressure conditions, the contents of each cell were drawn up into the sampling capillary and flash frozen at -80°C. The collected sample volumes were very small as a consequence of sampling single cells (<low μL). The cellular contents were then sprayed directly from the sampling capillary into a high resolution, high mass accuracy Orbitrap mass spectrometer using a static nanospray ionisation source. This technique provides a means of analysis that uses a soft ionisation technique to convey analytes into the mass spectrometer, minimising fragmentation of constituent molecules, making it suitable for analysis of complex biological samples. The mass spectra from each sample were then screened for the parent compounds and potential pharmaceutical metabolites and conjugates.
Summary of findings
For the first time, this study demonstrates the detection of exogenously applied pharmaceutical compounds in the contents of live, single plant cells using SCMS. Following exposure via saturated hydroponic solutions, four of the five studied pharmaceutical compounds (levetiracetam, metronidazole, clarithromycin and lincomycin) were detected in parent form. These compounds are thought to be translocated to other tissues via the symplastic pathway following root adsorption which allows them to cross cell membranes and thus explains their detection (7). Sulfadiazine, however, was not detected. This is likely because sulfadiazine is thought to follow the intercellular apoplastic pathway which prevents it from traversing the cell wall (7). This lack of detection thus highlights the efficacy of this technique for identifying the distribution of xenobiotic compounds in plant tissues.
Alongside these parent compounds, two transformation products of metronidazole and eight transformation products of clarithromycin were identified including two biologically active metabolites of clarithromycin that have not been previously documented in plant uptake studies.
These findings confirm the suitability of this application of SCMS for sampling and analysis of the contents of single cells in vivo. The work demonstrates accumulation and transformation of exogenously applied antibiotics in single cells in detectable concentrations for the first time and could have important implications for research into the ultimate chemical fate of xenobiotic compounds in environmental settings by helping to elucidate possible mechanisms and pathways for fixation and detoxification of xenobiotic compounds in plants.
For the first time, this study demonstrates the detection of exogenously applied pharmaceutical compounds in the contents of live, single plant cells using SCMS. Following exposure via saturated hydroponic solutions, four of the five studied pharmaceutical compounds (levetiracetam, metronidazole, clarithromycin and lincomycin) were detected in parent form. These compounds are thought to be translocated to other tissues via the symplastic pathway following root adsorption which allows them to cross cell membranes and thus explains their detection (7). Sulfadiazine, however, was not detected. This is likely because sulfadiazine is thought to follow the intercellular apoplastic pathway which prevents it from traversing the cell wall (7). This lack of detection thus highlights the efficacy of this technique for identifying the distribution of xenobiotic compounds in plant tissues.
Alongside these parent compounds, two transformation products of metronidazole and eight transformation products of clarithromycin were identified including two biologically active metabolites of clarithromycin that have not been previously documented in plant uptake studies.
These findings confirm the suitability of this application of SCMS for sampling and analysis of the contents of single cells in vivo. The work demonstrates accumulation and transformation of exogenously applied antibiotics in single cells in detectable concentrations for the first time and could have important implications for research into the ultimate chemical fate of xenobiotic compounds in environmental settings by helping to elucidate possible mechanisms and pathways for fixation and detoxification of xenobiotic compounds in plants.
References
1. X. Wu, L. K. Dodgen, J. L. Conkle, J. Gan. Sci. Total Environ. 536, 655-666 (2015).
2. T. Malchi, Y. Maor, G. Tadmor, M. Shenker, B. Chefetz. Environ. Sci. Technol. 48 (16), 9325-9333 (2014).
3. O. Paltiel, G. Fedorova, G. Tadmor, G. Kleinstern, Y. Maor, B. Chefetz. Environ. Sci. Technol. 50 (8), 4476-4482 (2016).
4. O. Guillaume-Gentil, T. Rey, P. Kiefer, A. J. Ibáñez, R. Steinhoff, R. Brönnimann, L. Dorwling-Carter, T. Zambelli, R. Zenobi, J. A. Vorholt. Anal. Chem. 89 (9), 5017-5023 (2017).
5. T. Fujii, S. Matsuda, M. Lorenzo Tejedor, T. Esaki, I, Sakane, H. Mizuno, N. Tsuyama, T. Masujima. Nat. Protoc. 10, 1445-1456 (2015).
6. N. Hanna, P. Sun, Q. Sun, X. Li, X. Yang, X. Ji, H. Zou, J. Ottoson, L. E. Nilsson, B. Berglund, O. J. Dyar, A. J. Tamhankar, C. Stålsby Lundborg. Environ. Int. 114, 131-142 (2018).
7. Y.-H. Chuang, C.-H. Liu, J. B. Sallach, R. Hammerschmidt, W. Zhang, S. A. Boyd, H. Li. Environ. Int. 131, 104976 (2019).
Further reading
Pharmaceuticals in the Environment (Issues in Environmental Science and Technology), R. E. Hester and R. M. Harrison (eds.), Royal Society of Chemistry, Cambridge, 2015 ISBN 978-1-78262-189-8
https://doi.org/10.1039/9781782622345
1. X. Wu, L. K. Dodgen, J. L. Conkle, J. Gan. Sci. Total Environ. 536, 655-666 (2015).
2. T. Malchi, Y. Maor, G. Tadmor, M. Shenker, B. Chefetz. Environ. Sci. Technol. 48 (16), 9325-9333 (2014).
3. O. Paltiel, G. Fedorova, G. Tadmor, G. Kleinstern, Y. Maor, B. Chefetz. Environ. Sci. Technol. 50 (8), 4476-4482 (2016).
4. O. Guillaume-Gentil, T. Rey, P. Kiefer, A. J. Ibáñez, R. Steinhoff, R. Brönnimann, L. Dorwling-Carter, T. Zambelli, R. Zenobi, J. A. Vorholt. Anal. Chem. 89 (9), 5017-5023 (2017).
5. T. Fujii, S. Matsuda, M. Lorenzo Tejedor, T. Esaki, I, Sakane, H. Mizuno, N. Tsuyama, T. Masujima. Nat. Protoc. 10, 1445-1456 (2015).
6. N. Hanna, P. Sun, Q. Sun, X. Li, X. Yang, X. Ji, H. Zou, J. Ottoson, L. E. Nilsson, B. Berglund, O. J. Dyar, A. J. Tamhankar, C. Stålsby Lundborg. Environ. Int. 114, 131-142 (2018).
7. Y.-H. Chuang, C.-H. Liu, J. B. Sallach, R. Hammerschmidt, W. Zhang, S. A. Boyd, H. Li. Environ. Int. 131, 104976 (2019).
Further reading
Pharmaceuticals in the Environment (Issues in Environmental Science and Technology), R. E. Hester and R. M. Harrison (eds.), Royal Society of Chemistry, Cambridge, 2015 ISBN 978-1-78262-189-8
https://doi.org/10.1039/9781782622345